REFERENCES:Previous Next Home
The sulfide hydrogenase from Pyrococcus furiosus contains a novel Asp-liganded FeS cluster [1]
Sulfide dehydrogenase is a versatile flavo-iron-sulfur protein that catalyzes both the reduction of polysulfide by NADPH and of NADP+ by ferredoxin [2]. Reduction of NADP+ has been proposed to be a necessary step in the disposal of reducing equivalents as H2. The physiological role of sulfur reduction, however, is not clear.
A detailed study of the prosthetic groups of the sulfide dehydrogenase through sequence analysis and EPR-monitored redox titrations has provided a clearer picture of the composition of sulfide dehydrogenases [1, my PhD thesis].The protein was shown to carry three [Fe-S] clusters, each of them atypical in some way: a putative [2Fe-2S] cluster with novel Asp(Cys)3 coordination and with the combination of physico-chemical properties hitherto exclusively ascribed to Rieske-type clusters; an apparently regular [3Fe-4S] cluster with an unusually high reduction potential; and a [4Fe-4S] cluster with unusual relaxation properties and reduction properties. The redox potential of the flavin is consistent with the proposed functions as S0-reductase and ferredoxin:NADP+ oxidoreductase, but the role of the high potential [2Fe-2S] and [3Fe-4S] in these reactions is not obvious.
Each of the subunits of SuDH appears to be a member of a different class of iron-sulfur flavoproteins. The SudB subunit is very similar to the reductase+ferredoxin systems from plants and algae. The SudA subunit carries the basic pattern found in a small class of iron-sulfur flavoproteins with iron-sulfur clusters of unusual properties, and for which crystallographic data are not yet available. It is likely that structural and functional studies of these homologous proteins may shed light on the mechanism of action and physiological functions of sulfide dehydrogenase.
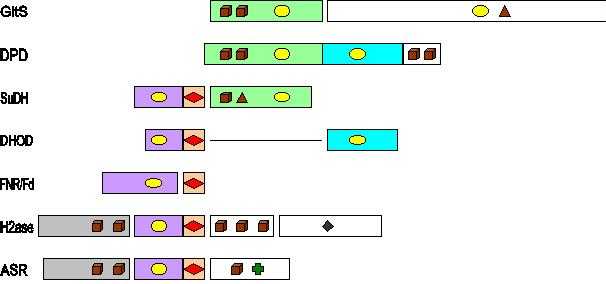
GltS, glutamate synthase (Azospirillum brasilense); DPD, dihydropyrimidine dehydrogenase (porcine liver); SuDH, sulfide dehydrogenase (P. furiosus); DHOD, dihydroorotate dehydrogenase (Lactococcus lactis); FNR, ferredoxin: NADP+ oxidoreductase (Spinach); Fd, ferredoxin (Spinach); H2ase, sulfhydrogenase (P. furiosus); ASR, assimilatory sulfite reductase (Salmonella typhimurium). Brown cubes stand for [4Fe-4S], brown triangles for [3Fe-4S], red diamonds for [2Fe-2S], yellow ellipses for FAD/FMN, a dark green diamond for the Ni-Fe dimer in hydrogenase, and a green cross for the siroheme in sulfite reductase.
[1]Novel
structure and redox chemistry of the prosthetic groups of the iron-sulfur
flavoprotein sulfide dehydrogenase from Pyrococcus furiosus; evidence
for a [2Fe-2S] cluster with Asp(Cys)3 ligands. W.R.
Hagen, P.J. Silva, M.A. Amorim, P.-L. Hagedoorn, H. Wassink, H. Haaker
and F.T. Robb. Journal of Biological Inorganic Chemistry (2000)
5, 527-534.
[2] Hydrogen production
from pyruvate by enzymes purified from the hyperthermophilic archaeon,
Pyrococcus
furiosus: A key role for NADPH.K. Ma, Z.H. Zhou and M.W.W.
Adams. FEMS Microbiol. Lett. (1994) 122: 245-250.